 |
display_and_projection notebook displays the wrong slice after backprojection.
Problem is that slice_num is set to central image slice which is half of 15, then reset to central sinogram BUT there are only 11 of those so slice_num becomes 5. This 5 is then used to display the result of the backprojection and you get
a slice that does not correspond to that displayed at the beginning.
The displayed help for AcquisitionData says that as_array returns a 3D array with first dimension sinograms, but I think this should say 4D with 1st dim TOF?
David
From: Kris Thielemans <[log in to unmask]>
Date: Saturday, 13 April 2019 at 13:11
To: David Atkinson <[log in to unmask]>, CCP <[log in to unmask]>
Subject: RE: VM
MAPEM is a known “problem”. Part of the exercise is to fix it!
From: Atkinson, David
Sent: 13 April 2019 13:08
To: Thielemans, Kris <[log in to unmask]>; [log in to unmask]
Subject: Re: VM
I think it downloaded all the PET stuff because it wasn’t super quick and it asked if I wanted to overwrite files.
Another problem?:
In MAPEM, I’ve just blasted through the cells without thinking, but I got some warnings from cell 17 (pasted below) and the image looks bad:
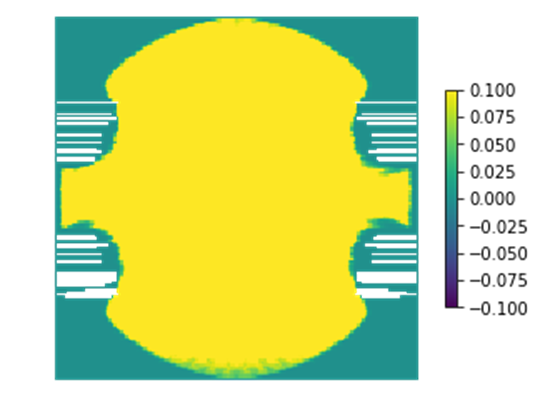
/usr/local/lib/python2.7/dist-packages/ipykernel_launcher.py:2: RuntimeWarning: invalid value encountered in divide
1
2
3
4
/usr/local/lib/python2.7/dist-packages/ipykernel_launcher.py:2: RuntimeWarning: divide by zero encountered in divide
/usr/local/lib/python2.7/dist-packages/ipykernel_launcher.py:3: RuntimeWarning: invalid value encountered in multiply
This is separate from the ipykernel package so we can avoid doing imports until
5
/usr/local/lib/python2.7/dist-packages/ipykernel_launcher.py:8: RuntimeWarning: invalid value encountered in double_scalars
/usr/local/lib/python2.7/dist-packages/ipykernel_launcher.py:2: RuntimeWarning: invalid value encountered in add
6
7
8
9
10
11
12
13
14
15
16
In [18]:
From: Kris Thielemans <[log in to unmask]>
Date: Saturday, 13 April 2019 at 12:57
To: David Atkinson <[log in to unmask]>, CCP <[log in to unmask]>
Subject: RE: VM
Hi David
It’s possible that the link wasn’t there when I made the VM. Did the download_PET script finish very quickly (i.e. did it actually download anything)? If so, we can just tell them to execute the download scripts (in fact, we’re telling
them already!)
Kris
PS: Ben just discovered a problem with notebook validation which you might run into when updating the VM. Trying to fix that now. sorry
From: Atkinson, David
Sent: 13 April 2019 12:53
To: Thielemans, Kris <[log in to unmask]>;
[log in to unmask]
Subject: Re: VM
The 4.9Gb VM contains all the data?
I had trouble running the PET reconstruct_measured_dat notebook with the file 20170809_NEMA_60min_UCL.l.hdr
I thought it was a repeat of the earlier issue with multiple file accesses. However, after running the PET data download script (which I had not done before as I assumed no need), it works. I think it’s because the download script creates
a symbolic link, which I assume is missing in the 4.9Gb VM. (It could also be because I have only just run updateVM for the first time).
From: CCP <[log in to unmask]> on behalf of Kris Thielemans <[log in to unmask]>
Reply-To: Kris Thielemans <[log in to unmask]>
Date: Saturday, 13 April 2019 at 00:47
To: CCP <[log in to unmask]>
Subject: Re: VM
Resent-From: David Atkinson <[log in to unmask]>
Done. (and some more doc re-org)
From: CCP-PETMR Developers list <[log in to unmask]>
On Behalf Of Thielemans, Kris
Sent: 12 April 2019 23:02
To: [log in to unmask]
Subject: Re: VM
Hi David
This is due to a known (but not well-documented) issue:
https://github.com/CCPPETMR/SIRF/issues/290
Essentially what happens is that you have one notebook—kernel reading the data, then the second notebook-kernel trying to read the same data and it cannot.
The only work-around is to use “File->Close and halt” after you’ve finished with a notebook (or just “Kernel->Shutdown”). I’ll add that the README. Johannes, you will have to warn people about his (so I’ll add it to the DocForInstructors).
Kris
From: Atkinson, David
Sent: 12 April 2019 22:54
To: Thielemans, Kris <[log in to unmask]>;
[log in to unmask]
Subject: VM
VM downloads page still needs correct VB version inserting.
Following the link, user gets a choice of two VMs. I chose the larger (4.9Gb) and it does seem to have the data pre-installed. This is good – only took 10minutes to download the whole lot over BT broadband.
In the MR notebooks, the first ran OK. Second gives the intentional error in the 2nd cell, BUT, the 3rd cell gives the message below. I’m assuming I wasn’t supposed to do something else as part of the exercise? Strange
thing is that this same h5 file is used OK in the first notebook.
File: /home/sirfuser/devel/install/python/sirf/Gadgetron.py
Line: 747
check_status found the following message sent from the engine:
---------------------------------------------------------------------------
error Traceback (most recent call last)
<ipython-input-3-239b343e6ff1> in <module>()
33
34
---> 35 acq_data = pMR.AcquisitionData('ptb_resolutionphantom_fully_ismrmrd.h5')
36
37 kdim = acq_data.dimensions()
/home/sirfuser/devel/install/python/sirf/Gadgetron.pyc in __init__(self, file)
745 if file is not None:
746 self.handle = pygadgetron.cGT_ISMRMRDAcquisitionsFromFile(file)
--> 747 check_status(self.handle)
748 def __del__(self):
749 if self.handle is not None:
/home/sirfuser/devel/install/python/sirf/Utilities.pyc in check_status(handle, stack)
363 repr(line) + ' of ' + file + '\n' + \
364 'the reconstruction engine output may provide more information'
--> 365 raise error(errorMsg)
366
367
error: ??? '"ISMRMRD File Error in ismrmrd_open_dataset (/home/sirfuser/devel/buildVM/sources/ISMRMRD/libsrc/dataset.c:969: Failed to open file.\\nISMRMRD HDF5 Error in H5Fopen (../../../src/H5F.c:579: unable to open file\\nISMRMRD HDF5
Error in H5F_open (../../../src/H5Fint.c:1168: unable to lock the file or initialize file structure\\nISMRMRD HDF5 Error in H5FD_lock (../../../src/H5FD.c:1821: driver lock request failed\\nISMRMRD HDF5 Error in H5FD_sec2_lock (../../../src/H5FDsec2.c:939:
unable to flock file, errno = 11, error message = \'Resource temporarily unavailable\'" exception caught at line 490 of /home/sirfuser/devel/buildVM/sources/SIRF/src/xGadgetron/cGadgetron/cgadgetron.cpp\nthe reconstruction engine output may provide more information'
From: Kris Thielemans <[log in to unmask]>
Date: Friday, 12 April 2019 at 18:19
To: David Atkinson <[log in to unmask]>, CCP <[log in to unmask]>
Subject: RE: {SPAM?} VB
Good catch. The downloads page should say VB 5.2.26 for v2.0.0! Edo/Evgueni?
We could change the VM INSTALL.md to point to the version of old VBs I guess.
From: CCP-PETMR Developers list <[log in to unmask]>
On Behalf Of Atkinson, David
Sent: 12 April 2019 18:16
To: [log in to unmask]
Subject: {SPAM?} VB
Do you want people to use VB 5.2.12 as stated on
http://www.ccppetmr.ac.uk/downloads ?
If so, then at point 2 on Wiki page
https://github.com/CCPPETMR/CCPPETMR_VM/blob/master/INSTALL.md
we should unlink the “VirtualBox” as this link takes you to a page with a massive button for downloading VB 6.0
David
From: CCP <[log in to unmask]> on behalf of Casper da Costa-Luis <[log in to unmask]>
Reply-To: Casper da Costa-Luis <[log in to unmask]>
Date: Friday, 12 April 2019 at 17:51
To: CCP <[log in to unmask]>
Subject: Re: Jupyter notebooks from your/our laptops?
Resent-From: David Atkinson <[log in to unmask]>
Yes, you may have to expose the port/enable network access in your computer's firewall (if you have one).
On Fri, 12 Apr 2019 at 17:42, Thielemans, Kris <[log in to unmask]> wrote:
What I don’t understand about this is how people can connect to this port. It will be blocked by firewalls etc etc…
From: Casper da Costa-Luis <[log in to unmask]>
Sent: 12 April 2019 16:13
To: Brown, Richard <[log in to unmask]>
Cc: Thomas, Ben <[log in to unmask]>; Thielemans, Kris <[log in to unmask]>; [log in to unmask]
Subject: Re: Jupyter notebooks from your/our laptops?
Successfully got this working (from within a docker container but that's besides the point):
for JUPYTER_PORT in $(seq 8890 8899); do
[ -d ~/SIRF-Exercises-$JUPYTER_PORT ] || cp -r /devel/SIRF-Exercises ~/SIRF-Exercises-$JUPYTER_PORT
jupyter notebook --ip 0.0.0.0 --port $JUPYTER_PORT --no-browser --notebook-dir ~/SIRF-Exercises-$JUPYTER_PORT &
donejupyter notebook list
If people accidentally kill a jupyter server, you can just rerun the above safely (it won't affect things which are already started and won't overwrite old working directories).
Best,
Casper
On Fri, 12 Apr 2019 at 16:01, Brown, Richard <[log in to unmask]> wrote:
No need for other cloud operators I think. Your suggestion at the end looks extremely straightforward, so worse case scenario we should be able to get by like that (a few from yours, mine, Kris’, Johannes’ etc.). I would expect at least half (28 total) to be using the VM, so it should be doable.
Anyway, hopefully it won’t be necessary, just keeping all the doors open.
Cheers,
Richard
On 12 Apr 2019, at 13:28, Casper da Costa-Luis <[log in to unmask]> wrote:
Hi Richard,
TL;DR: This email finishes with a simple suggestion.
Happy to still do this, though I'll be arriving early Sunday morning so don't know if I'll be there for the start of the session.
We could set this up on another cloud provider if you want (AWS docker, Google Cloud docker, etc...).
To do it on your own laptop, I've included instructions below based on https://github.com/CCPPETMR/SIRF-SuperBuild/tree/master/docker#more-information.
Assuming you have docker installed (easiest on ubuntu, https://docs.docker.com/install/linux/docker-ce/ubuntu/):
sudo apt-get update
sudo apt-get install apt-transport-https ca-certificates curl gnupg-agent software-properties-common
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add -
sudo add-apt-repository "deb [arch=amd64] https://download.docker.com/linux/ubuntu $(lsb_release -cs) stable"
sudo apt-get update
sudo apt-get install docker-ce docker-ce-cli containerd.io docker-compose
Then you need:
git clone https://github.com/CCPPETMR/SIRF-SuperBuild/
cd SIRF-SuperBuild/docker
sed -i -r 's/[0-9]+://' docker-compose.srv.yml
sed -i -r 's/container_name:\s+sirf/#/' docker-compose.yml
./sirf-compose-server up -d --scale sirf=3 sirf # start 3 servers
./sirf-compose-server ps # print out exposed ports
This will give you a <port> for each server:
... 0.0.0.0:32783->8888 ...
Meaning point a browser to localhost:32783 (password virtual).
If you connect your laptop to a network/make it a hotspot you can find out its <IP>
(via ifconfig) so that other users on the network can connect to <IP>:<port>
./sirf-compose-server stop # stop all servers
Note that this shares the "./devel" folder as the jupyter root for all servers!
You probably want some isolation.
I was planning on making a simple script for this.However, in light of the suggestion below, though, you may not need it.
TL;DR
If you have SIRF & jupyter just natively installed on any machine, couldn't you simply:
- gadgetron >& /dev/null &
- clone SIRF-Exercises
- make a few copies of the folder
- for folder number <n> in folders: jupyter notebook --ip 0.0.0.0 --port 888<n> --no-browser &
Best,
Casper
On Fri, 12 Apr 2019 at 10:36, Brown, Richard <[log in to unmask]> wrote:
Hi Casper,
Not sure which email address you prefer me to use so sending to both.
We’re having a few troubles with Azure - Are you still ok for people to remotely connect to your laptop to run their Jupyter notebooks?
Would it be possible for more of us to set this up? If so, could you let us know what we need to do?
Cheers,
Richard
Research Associate in Medical Imaging Physics at University College London,
Institute of Nuclear Medicine, UCL Hospital, Tower 5,
235 Euston Road, London NW1 2BU, UK
To unsubscribe from the CCP-PETMR-DEVEL list, click the following link:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?SUBED1=CCP-PETMR-DEVEL&A=1
To unsubscribe from the CCP-PETMR-DEVEL list, click the following link:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?SUBED1=CCP-PETMR-DEVEL&A=1
To unsubscribe from the CCP-PETMR-DEVEL list, click the following link:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?SUBED1=CCP-PETMR-DEVEL&A=1
To unsubscribe from the CCP-PETMR-DEVEL list, click the following link:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?SUBED1=CCP-PETMR-DEVEL&A=1
To unsubscribe from the CCP-PETMR-DEVEL list, click the following link:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?SUBED1=CCP-PETMR-DEVEL&A=1
To unsubscribe from the CCP-PETMR-DEVEL list, click the following link:
https://www.jiscmail.ac.uk/cgi-bin/webadmin?SUBED1=CCP-PETMR-DEVEL&A=1