On Nov 10, 2014, at 11:20 AM, Bryson Dietz <[log in to unmask]> wrote:
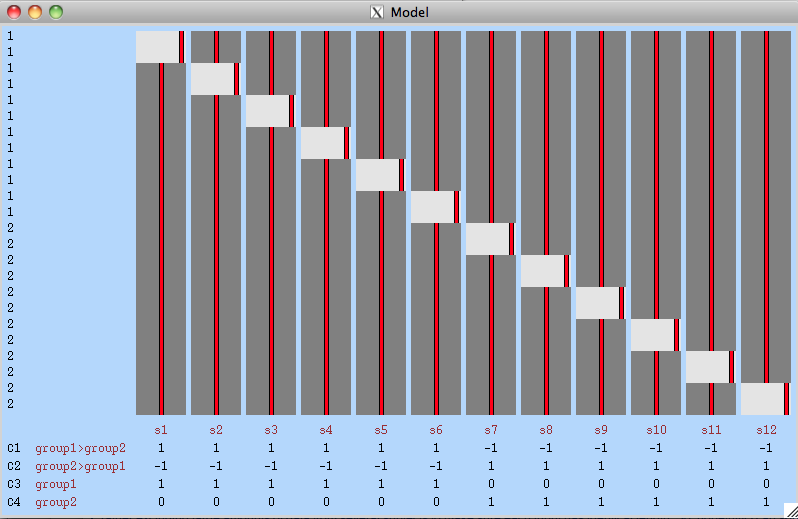
Hello Eugene and David,Here is my analysis, to be sure everything I am doing makes sense:1) Use MELODIC on each run for each subject (in each group) to remove unwanted noise2) Apply a group concat-ICA on the denoised data from both groups (registered to standard space)3) Feed the Melodic_IC output from step 2 into Dual-Regression (simplified GLM shown below)
<image.png>
4) Use the method mentioned by David to average the runs (dr_stage2_output) for each subject.5) Run randomize on each componentI guess I would not need a GLM for randomize, since I only have one input (i.e. dr_stage2_avg_ic0004).Will this method show me statically significant differences between groups for each IC (that I run randomize on)?Thanks again for taking the time to help me out here,BrysonOn Mon, Nov 10, 2014 at 10:41 AM, Eugene Duff <[log in to unmask]> wrote:Hi -I had missed you were using randomise: this makes the multi-level approach tricky, but the model you suggested above should work (ensuring you set exchangability blocks as subjects). This will be equivalent to taking the means.Cheers,EugeneOn 9 November 2014 23:07, Bryson Dietz <[log in to unmask]> wrote:Great, thanks Eugene and David. I appreciate your responses.
If I may be so BOLD (had to throw in a pun) as to ask a follow up question; are these two methods equivalent? Or is one more valid than the other?
Thanks again,
Bryson
Hi Bryson,Yes, you could average the dr_stage2 output from the runs/sessions. I posted some code to help with this procedure in this thread:JISCMail - FSL Archive - Re: NETWORK INTEGRATION SCORE (https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=FSL;6b8d55bf.1109)Hope this helps.Cheers,DavidOn Nov 8, 2014, at 2:53 PM, Bryson Dietz <[log in to unmask]> wrote:Hello,I just have a question regarding my resting-state analysis (3 runs, single-session, 6 subjects (for each group), 2 groups). Initially I made my GLM, essentially the same as mentioned below (I pasted the image from the link below in this email).Upon reading his response from Stephen Smith,
https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=ind1010&L=fsl&F=&S=&P=163366It appears that this analysis ignores variance between subjects. In the above link Stephen mentions "Hence I would recommend just averaging the 3 sessions' images (output by the dual regression) for each subject"I would just like to clarify, does Stephen mean to average the sessions (in my case runs) output from the dual-regression run from the above link (with the GLM shown below)? Also, I am assuming that I can simply exchange sessions with runs.I hope that I have made my question clear!Thanks,Bryson