 |
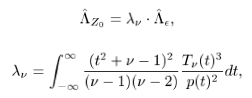
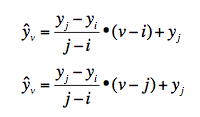
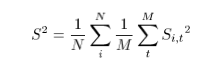
Right - yes, for (e.g.) the Z case it's set to 100 as anything larger than that gives virtually the same result.Cheers.
On 19 Jan 2012, at 20:56, YAN Chao-Gan wrote:Thanks, Steve!
Sorry for my unclear statement.
To my understanding, easythresh will call smoothest without passing the DOF parameter. However, in smoothest, DOF will default to 100 if nothing specified, is that right? What DOF should we use if we perform easythresh on a Z statistical image?
Best,
Chao-GanOn Thu, Jan 19, 2012 at 12:48 PM, Stephen Smith <[log in to unmask]> wrote:
I don't think it does - easythresh (at least my version) doesn't pass in a DoF.Cheers.On 18 Jan 2012, at 22:14, YAN Chao-Gan wrote:Dear FSL experts,
Recently I used easythresh on a Z stats map for multiple comparison correction.
I found easythresh will call smoothest with the degree of freedom set at the default value of 100. The DOF of statistical Z is not 100, right? Is there any reason to use 100 as DOF for Z stats map?
Thanks!
Chao-Gan
---------------------------------------------------------------------------
Stephen M. Smith, Professor of Biomedical Engineering
Associate Director, Oxford University FMRIB Centre
FMRIB, JR Hospital, Headington, Oxford OX3 9DU, UK
+44 (0) 1865 222726 (fax 222717)
[log in to unmask] http://www.fmrib.ox.ac.uk/~steve
---------------------------------------------------------------------------
---------------------------------------------------------------------------
Stephen M. Smith, Professor of Biomedical Engineering
Associate Director, Oxford University FMRIB Centre
FMRIB, JR Hospital, Headington, Oxford OX3 9DU, UK
+44 (0) 1865 222726 (fax 222717)
[log in to unmask] http://www.fmrib.ox.ac.uk/~steve
---------------------------------------------------------------------------